In forensic DNA analysis, one of the biggest challenges is dealing with degraded DNA—samples that have damaged over time. DNA consists of long fragments while in the cell, but when exposed to environmental factors like heat, moisture, chemicals, or microbial digestion these long fragments break into smaller fragments. To assess DNA degradation, forensic scientists often rely on the degradation index (DI). While DI is informative for Short Tandem Repeat (STR) testing and deciding if alternate methods should be pursued, it becomes irrelevant when applied to modern Single Nucleotide Polymorphism (SNP) testing, particularly on massively parallel DNA sequencing platforms.
The Origins and Development of the Degradation Index
As forensic DNA testing evolved in the 1990s and early 2000s, STR profiling became the dominant method for human identification. However, as samples collected from crime scenes were often degraded, forensic scientists needed a metric to evaluate whether the DNA was intact enough to generate a STR profile. The degradation index was developed to assess the integrity of DNA samples by comparing the amplification of targeted DNA fragments of different lengths.
The introduction of the Quantifiler® Human DNA Quantification Kit by Applied Biosystems (now Thermo Fisher Scientific) was pivotal. Released in the early 2000s, this kit became widely used for quantifying human DNA, determining if inhibitors were present in a sample and assessing degradation levels. The Quantifiler® kit, and later versions like the Quantifiler® Trio DNA Quantification Kit, measure DI by comparing the amplification of two DNA fragments: a shorter fragment (80 base pairs) and a longer fragment (214 base pairs).
A high DI indicates that the longer fragment has degraded significantly compared to the shorter one, suggesting that the DNA sample is degraded and may not be suitable for obtaining a full STR profile. Here is the how degradation index works:
-
DI < 1: Indicates intact DNA, suitable for standard STR testing.
-
DI between 1 and 10: Suggests moderate degradation. STR testing may still be possible, though the sample may yield a partial profile. Labs may also consider alternative methods like mini-STRs.
-
DI > 10 or undefined: Indicates significant degradation, indicating that STR testing may be unsuccessful and alternate approaches such as mini-STRs, low-template DNA testing, or mitochondrial DNA sequencing should be considered.
STR testing relies on analyzing repeated sections of DNA and because of constraints of capillary electrophoresis often require amplification of relatively long DNA fragments (usually up to 400 base pairs). DI informs forensic scientists about the degree of degradation and helps determine whether a sample is suitable for STR analysis.
Why DI is Irrelevant for DNA Sequencing
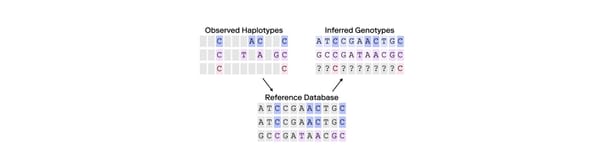
While DNA degradation also is an issue for DNA sequencing, DI is not a suitable metric for measuring it. DI is designed to evaluate degradation based on the amplification of longer DNA fragments than typically encountered with short fragment DNA sequencing. SNPs can be accurately measured from much shorter DNA fragments than required for STR typing.
For example, in cases of severe degradation, where no 200 base pair fragments are present, the degradation index would be undefined (division by zero), as it compares the amplification of a short fragment (80 base pairs) to a long fragment (214 base pairs). Even when the longer fragments are absent as indicated by the DI, SNP sequencing can still proceed successfully because the short fragments necessary for SNP analysis may remain sufficiently intact. Thus, in this context, DI is uninformative.
DI is an effective tool for STR testing, but it was never designed for DNA sequencing—particularly for SNP-based methods. As forensic testing evolves, it is crucial to recognize that not all tools are fit for every purpose.
Conclusion
At Othram, we pride ourselves on building tools that are purpose-built specifically for forensics and forensic SNP testing. The DI is not flawed—it simply was not intended for the parameters of DNA sequencing. Relying on tools designed for different purposes can lead to suboptimal outcomes in forensic genetic genealogy.
This issue is about responsible science, not for advocating for any one company’s internally accepted methods. Success in forensic genetic genealogy requires investing in purpose-built tools that are optimized for the unique challenges of this field. To drive real progress, we must build with intention, ensuring every tool is tailored to the task at hand.
If you are not ready to onboard this new technology in your own forensic setting yet, come to Othram. Our team operates the world's first purpose-built forensic laboratory for forensic genetic genealogy. We developed Forensic-Grade Genome Sequencing® or FGGS® to enable ultra-sensitive detection of distant relationships. It's part of our Multi Dimensional Forensic Intelligence (MDFI) platform.
More forensic genetic genealogy cases have been solved with Othram FGGS® than any other method. Let’s work together to unlock answers and bring justice to those who need it most. Get started here.