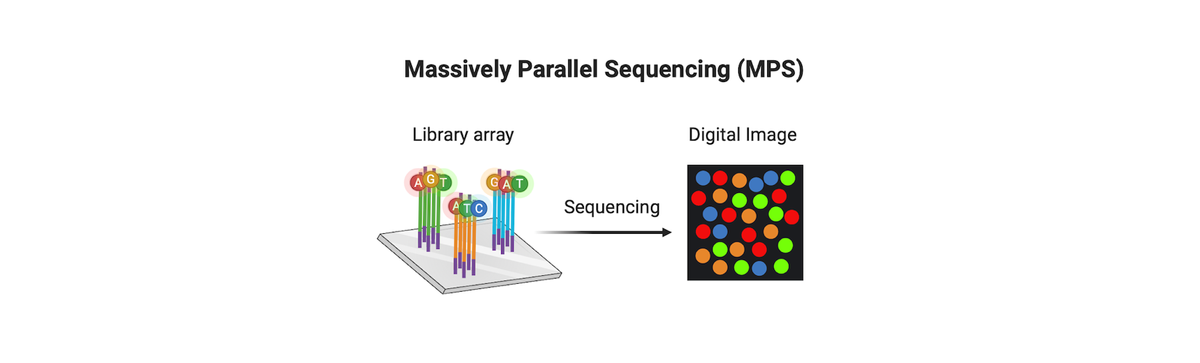
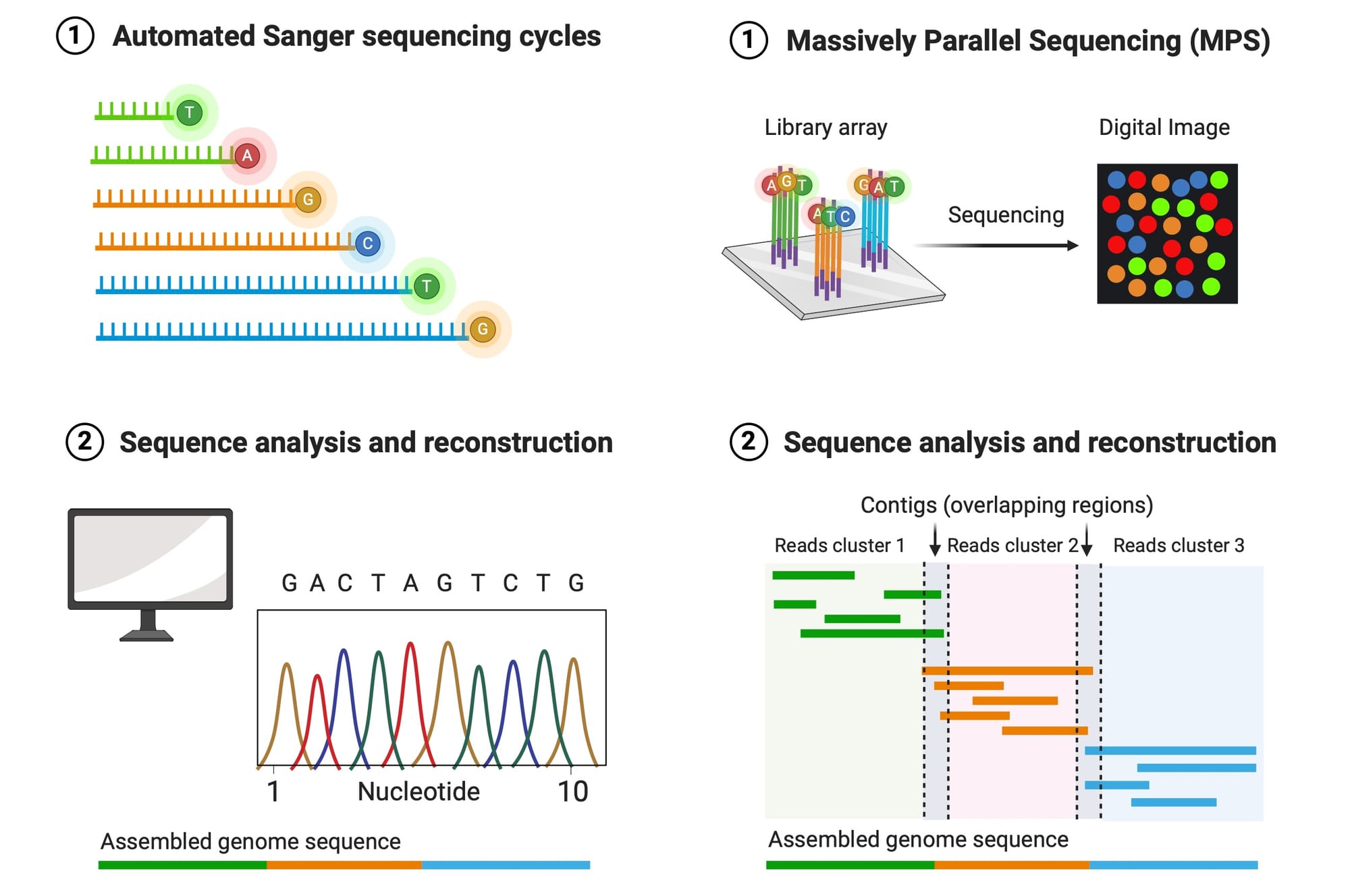
Massively parallel sequencing (MPS), whether for direct comparison, near kinship associations, or whole genome sequencing (WGS) for distant kinship associations, has become a transformative tool in forensic science. Of the various MPS-based technologies, WGS consistently yields better results from forensic evidence, particularly in cases involving low-quantity or highly degraded DNA samples. These advancements have made it possible to extract useful genetic information from cases that were previously considered unsolvable, bringing long-awaited resolution and justice to victims, families, and communities.
However, while MPS and single nucleotide polymorphisms (SNP) testing are powerful techniques, not every service provider claiming decades of forensic DNA testing experience is truly equipped for MPS. Experience with traditional forensic DNA analysis, such as STR typing, does not translate into expertise with MPS, which is a complex and nuanced technology. Similarly, those claiming expertise in genetic genealogy without a deep understanding of MPS and SNP testing may lack a foundational understanding of how to build SNP profiles. Inadequate, incomplete, or inaccurate DNA profiles can severely compromise the effectiveness of genetic genealogy, making it difficult or even impossible to resolve cases.
Othram, the first provider of SNP profiles purpose-built for forensic genetic genealogy, has been leading this field since 2018. The technology is new to the forensic field and claims of a decade or more of experience may be true but are not necessarily accurate. MPS is not just a simple upgrade from STR typing—it’s a fundamental shift in how forensic labs approach genetic analysis. As such, while more labs are beginning to adopt WGS, few so far are truly prepared to handle its complexities and potential.
Forensic DNA Experience Doesn't Ensure Success with MPS
As forensic stakeholders work toward integrating WGS, it’s essential to recognize that not all laboratories are equally ready. MPS/WGS technologies are fundamentally different from the capillary electrophoresis (CE) systems currently in use for STR testing. Thus, additional education, training, and comprehensive validation are critical steps that must be undertaken before a laboratory can effectively offer these services.
One common misconception is that experience with traditional forensic DNA analysis is sufficient to ensure success in implementing WGS. Relying solely on past experience is not enough. While forensic professionals may be skilled in handling DNA using CE and short tandem repeats (STR) markers, WGS introduces unique challenges that require specialized knowledge. Laboratories must address new technical hurdles, such as PCR duplicates, the quality of raw reads, and the effects of missing or low-quality data. Moreover, sequencing complex and GC-rich regions of the genome, handling homopolymers, and managing mapping and read errors all pose significant and new challenges that must be addressed.
Additionally, DNA extraction methods may need to be adapted, particularly for degraded samples that select for short fragments to improve typing success. Platform chemistries also come with their own set of challenges—some are more prone to issues with homopolymers, while others experience increased substitution errors. Factors such as the number of multiplexed samples and library preparation can impact sequencing depth and the quality of interpretation.
Software also plays a key role in analyzing the vast amount of data generated by MPS/WGS technologies. While these tools offer a streamlined approach from raw data to results, they can become problematic when used without sufficient knowledge and understanding. In a field just beginning to adopt WGS, forensic scientists must grasp the systems in use and avoid over-reliance on “black box” solutions. It’s not only the “black boxes” that are problematic; off-the-shelf solutions from other industries also get adopted in forensics, which may be unvalidated, with the blind hope that they’ll perform appropriately. This wishful belief can be equally detrimental. Lastly, relying on bioinformatics consultants to perform custom analyses can lead to results that are opaque and often irreproducible. These perceived approaches still require validation, testing and training. Forensic science deserves dedicated, purpose-built solutions that are transparent and objective, allowing methods to be refined and improved over time; in other words, robust, sustainable solutions should be sought. This tenet is essential to reduce errors similar to those that occurred during past technological transitions, such as mixture interpretation issues or problems introduced by data smoothing.
Lessons from Past Technological Shifts
The challenges of adopting MPS/WGS are not unique. The introduction of PCR and STR markers in the 1990s also brought about significant hurdles. Initially, restriction fragment length polymorphism (RFLP) typing was the dominant DNA analysis method, but it was eventually replaced by PCR-based systems due to PCR’s higher sensitivity and ability to analyze relatively degraded DNA. However, the transition to PCR required substantial adjustments. Laboratories had to address new concerns, such as contamination, robust chemistries, the validation of CE equipment, and the development of software for data interpretation. These challenges echo those being faced today with MPS and WGS.
Just as RFLP did not automatically grant knowledge needed for PCR-based methods adoption, CE-based experience will not fully prepare forensic professionals for MPS/WGS. New methods introduce new obstacles that must be recognized and addressed to ensure reliable results. Forensic scientists today need to engage with the technical demands of WGS, just as past generations did when they embraced PCR.
The Path Forward for MPS and WGS in Forensic Science
The foundations of forensic DNA are solid, developed over decades of investment in infrastructure, validation, and quality assurance. However, moving forward with MPS/WGS will require the same careful approaches that were used to integrate PCR and STRs into forensic practice. There are many topics specific to MPS/WGS that must be addressed, including PCR duplicates, PCR artifacts, reaction stringency, processing raw reads, sequencing complex regions, and managing missing or low-quality data. Additionally, extraction methods may need to be revisited to accommodate short fragments, particularly from degraded samples.
Implementing robust metrics is equally important when adopting MPS, as many of the quality indicators used for traditional DNA typing may not fully apply to MPS workflows. For instance, degradation index, which was critical for STR typing, is largely uninformative for SNP-based sequencing. Similarly, metrics like call rate and Q scores, which have been standard in other genomic applications, require further refinement and better understanding in the forensic context. This is why our research team dedicates so much time to writing about metrics.
Sequencing chemistries also present unique challenges, such as issues with homopolymers or increased substitution errors, depending on the platform. The number of multiplexed samples, library preparation quality, and sequencing depth all impact the success of an analysis. As WGS begins to be used more widely, the software that facilitates data analysis must be carefully evaluated to ensure it is functioning effectively and accurately. In the best case scenario, software and laboratory methods are optimized together, ensuring that you get the most from your forensic evidence and the data derived from it.
At Othram, we have spent years developing and advocating performance metrics, such as successful typing rates, minimal DNA consumption, kinship distance associations, error rates, and capability to perform genealogical research. In forensic genetic genealogy, the goal is to create ultra-sensitive DNA profiles capable of detecting distant genetic relationships. This requires getting lots of correct data from your forensic evidence and as little incorrect data as possible. It will often be the difference between a case solve and a DNA dead end.
If you are not ready to onboard this new technology in your own forensic setting yet, come to Othram. Our team operates the world's first purpose-built forensic laboratory for forensic genetic genealogy. We developed Forensic-Grade Genome Sequencing® or FGGS® to enable ultra-sensitive detection of distant relationships. It's part of our Multi Dimensional Forensic Intelligence (MDFI) platform.
More forensic genetic genealogy cases have been solved with Othram FGGS® than any other method. Let’s work together to unlock answers and bring justice to those who need it most. Get started here.