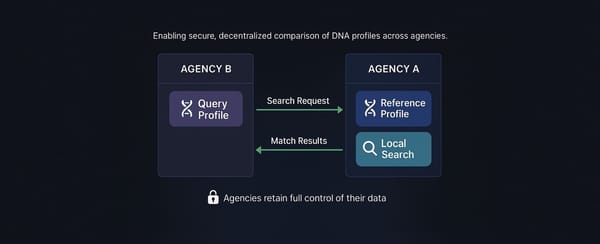
The Othram research team is building tools to greatly reduce the time and effort required by law enforcement and forensic genetic genealogists to resolve forensic cases. All these tools—like the one we’re discussing today—are part of Othram’s Multi-Dimensional Forensic Intelligence (MDFI) platform. You can learn more about the MDFI platform here.
In our last post, we introduced the “Shared Matches” view, which helps investigators and forensic genetic genealogists identify which DNA matches are directly connected to each other. By allowing you to see who shares DNA with both the searched profile and other matches, investigators are able to understand how individuals might be related on a more granular level. But this brings up an important question: with potentially hundreds of matches to analyze, how do you know which two matches to examine using the “Shared Matches” view?
This is where clustering comes into play. Clustering works at the macro level to organize DNA matches into distinct groups or clusters based on shared DNA segments. These clusters often correspond to different branches of a family tree, providing insight into how different individuals are related to both the searched profile (the person being analyzed) and each other. By grouping matches in this way, investigators can more easily identify potential relatives and trace family lines. By using clustering to identify which groups of matches belong together, you can more effectively decide which matches to analyze further with the shared matches feature, ultimately streamlining and focusing your forensic investigations.
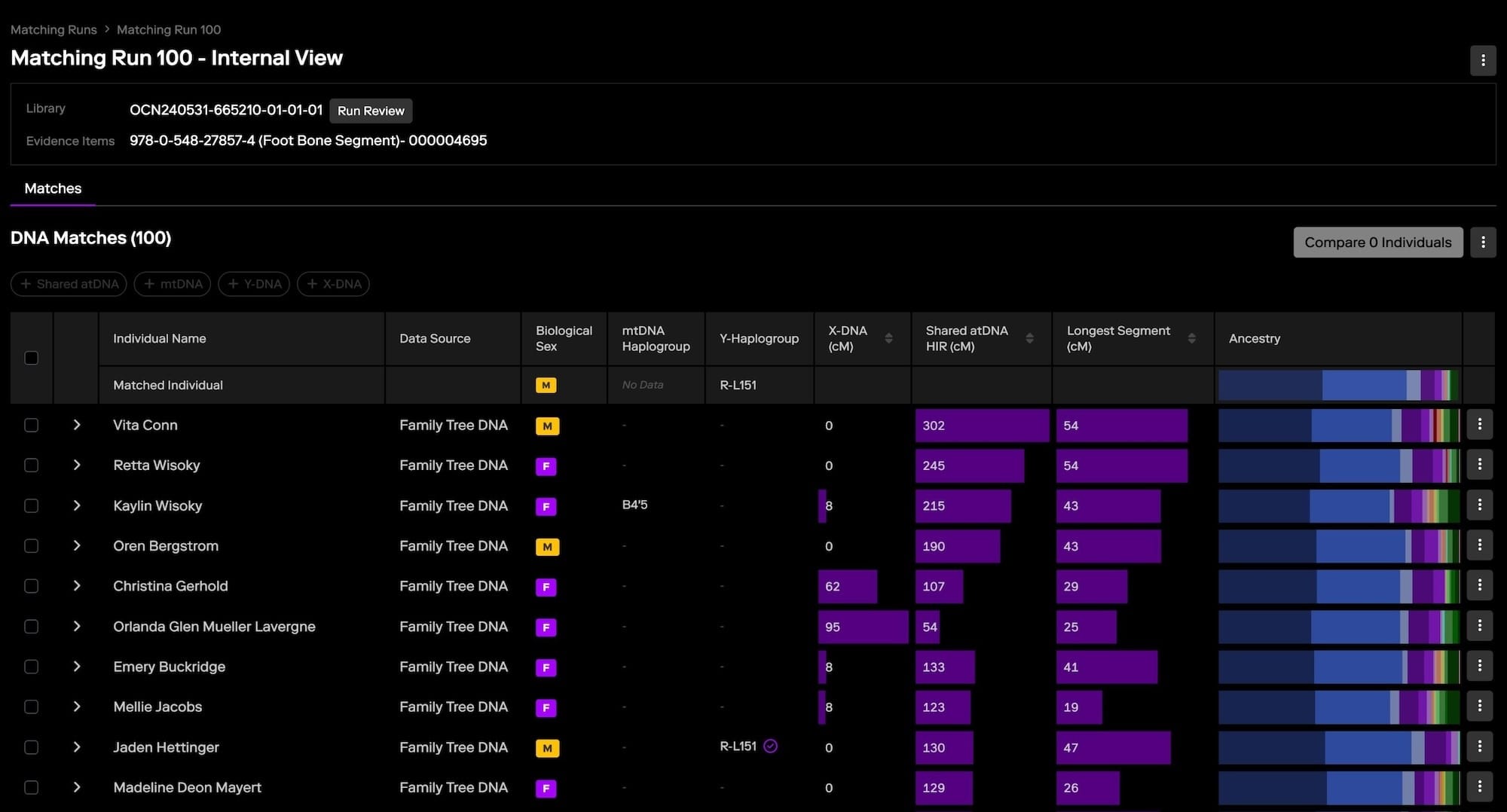
How Clustering Works
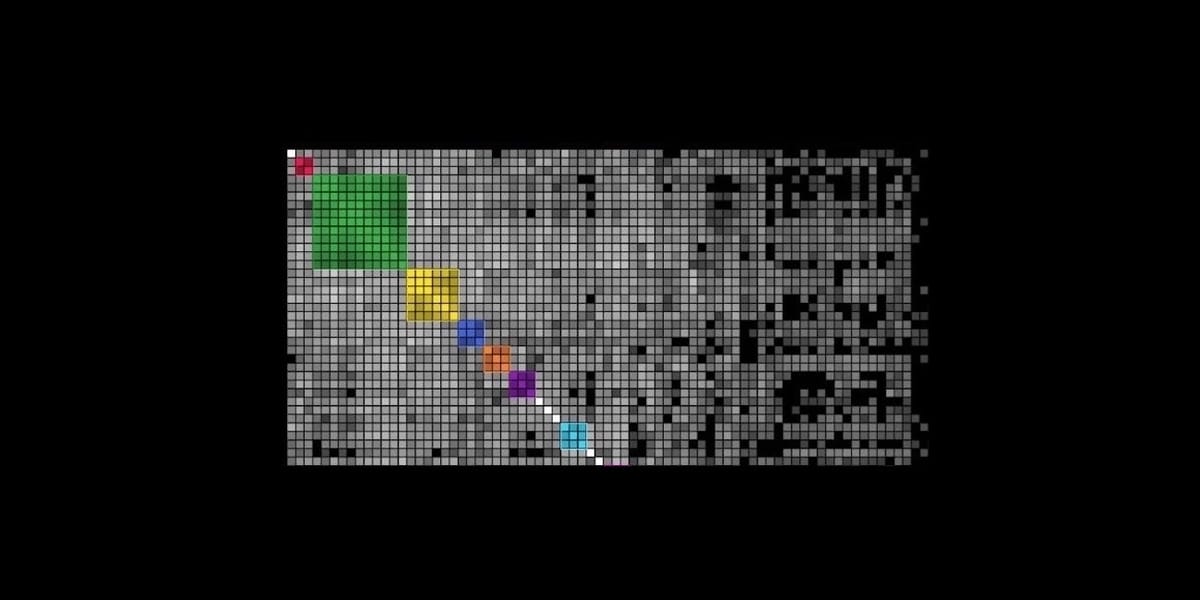
Clustering in genetic genealogy is a method that organizes DNA matches into distinct groups based on shared segments of DNA. These clusters often correspond to specific branches of a family tree, providing a clearer picture of how different matches are related to the searched profile and each other. The process begins with identifying shared DNA segments between the searched profile—the individual being analyzed—and their matches within the genetic genealogy database. Once these shared segments are identified, the matches are grouped into clusters based on the amount and location of shared DNA.
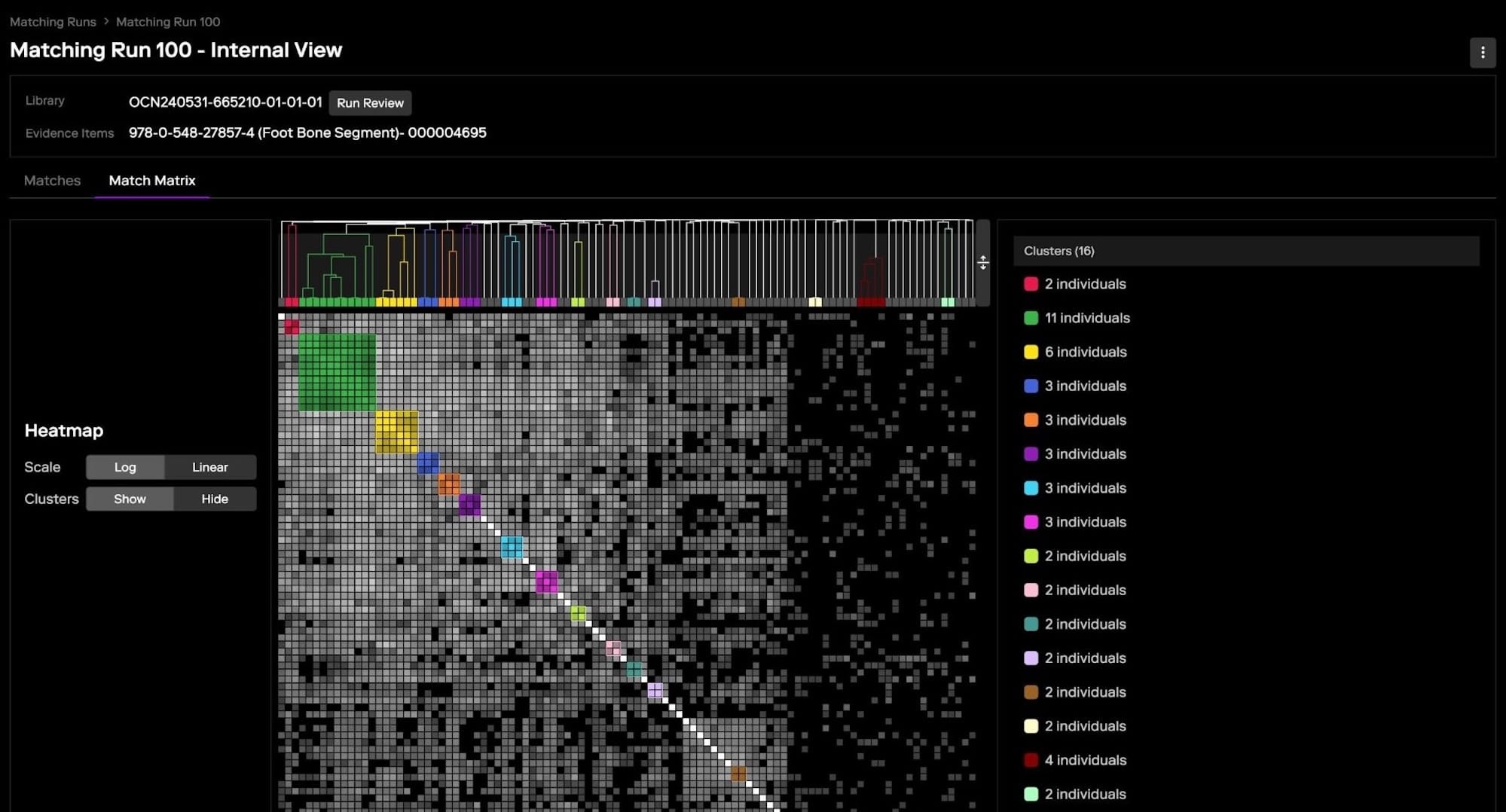
These clusters represent groups of individuals who likely share a common ancestor, making it easier to trace family connections and focus on relevant lines of descent. Clustering can often be visualized using various tools that create graphical representations, helping investigators quickly see how matches are connected and identify distinct clusters that align with different family branches.
Practical Applications in Forensic Investigations
Clustering is particularly useful in forensic investigations where the goal is to identify an unknown suspect or a victim. When a DNA profile is uploaded to the MDFI platform, the forensic search algorithm might generate a large list of matches. Analyzing each match individually can be time-consuming and overwhelming, especially if many of the matches are distant relatives. Clustering simplifies this process by grouping the matches into clusters that represent different family branches, allowing investigators to focus on the clusters that are most relevant to their case.
The quality of forensic DNA profiles can vary significantly from consumer DNA profiles. Forensic DNA profiles may lack certain information or markers, or even contain incorrect markers—due to the degraded nature of the DNA samples or the challenging conditions under which they are collected. This can lead to a higher presence of noise, where irrelevant matches clutter the results, making it harder to identify key connections.
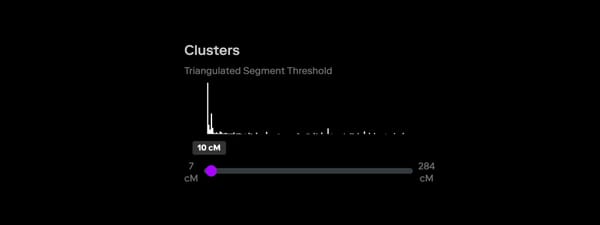
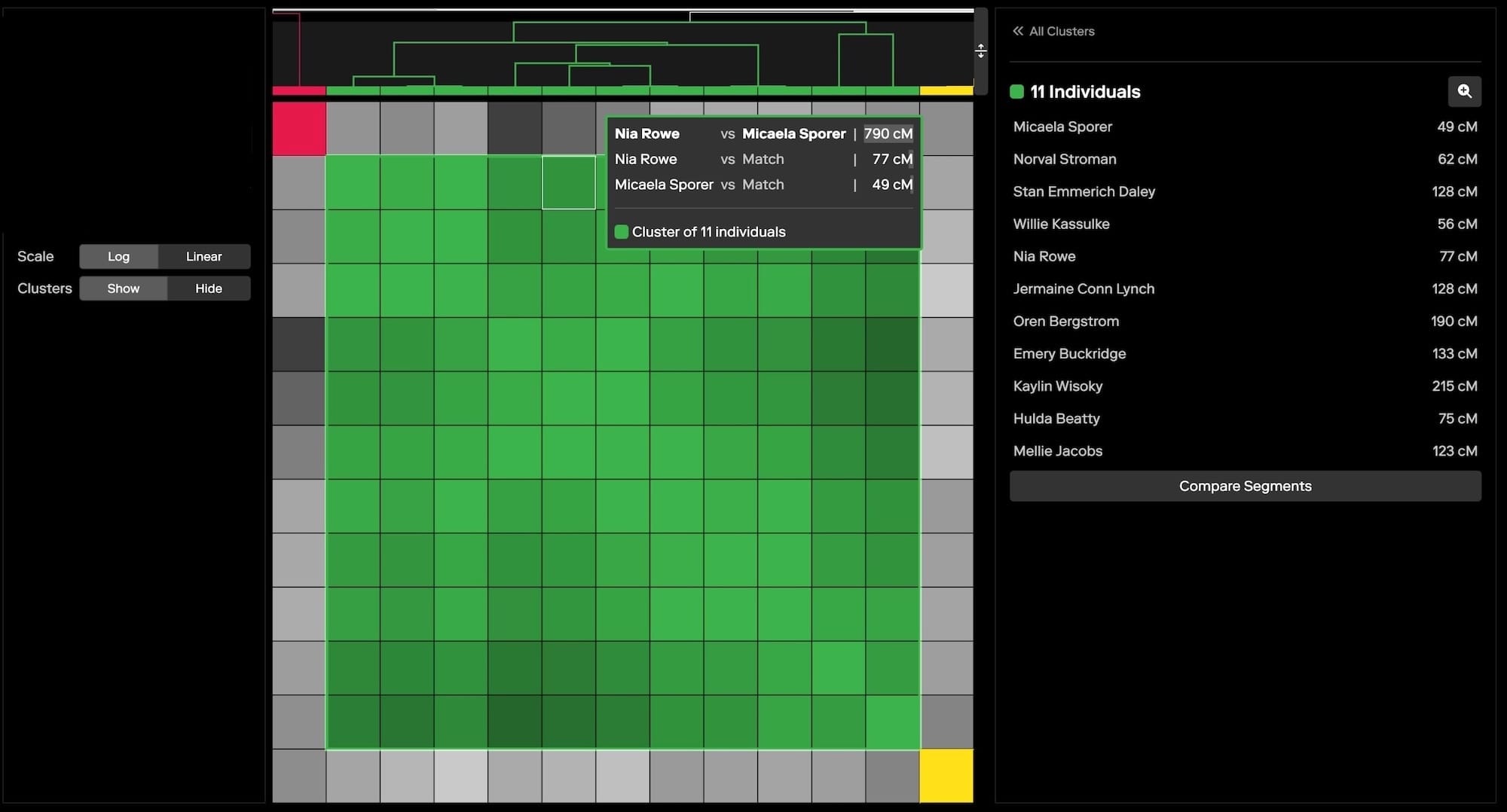
Clustering and the “Shared Matches” view are crucial tools in these situations. Clustering helps to distinguish relevant matches from this noise by organizing matches into meaningful groups that likely represent true genetic relationships. This is particularly valuable in forensic investigations, where fewer matches are typically available compared to consumer searches. Because clustering works especially well with distant relatives, it can be instrumental in identifying connections that might otherwise be overlooked in cases where close relatives are not present in the database. You can further use the "Segment Comparison" view, shown below, to examine the amounts and locations of shared DNA amongst the clustered matches, as an additional metric for identifying relevant matches.
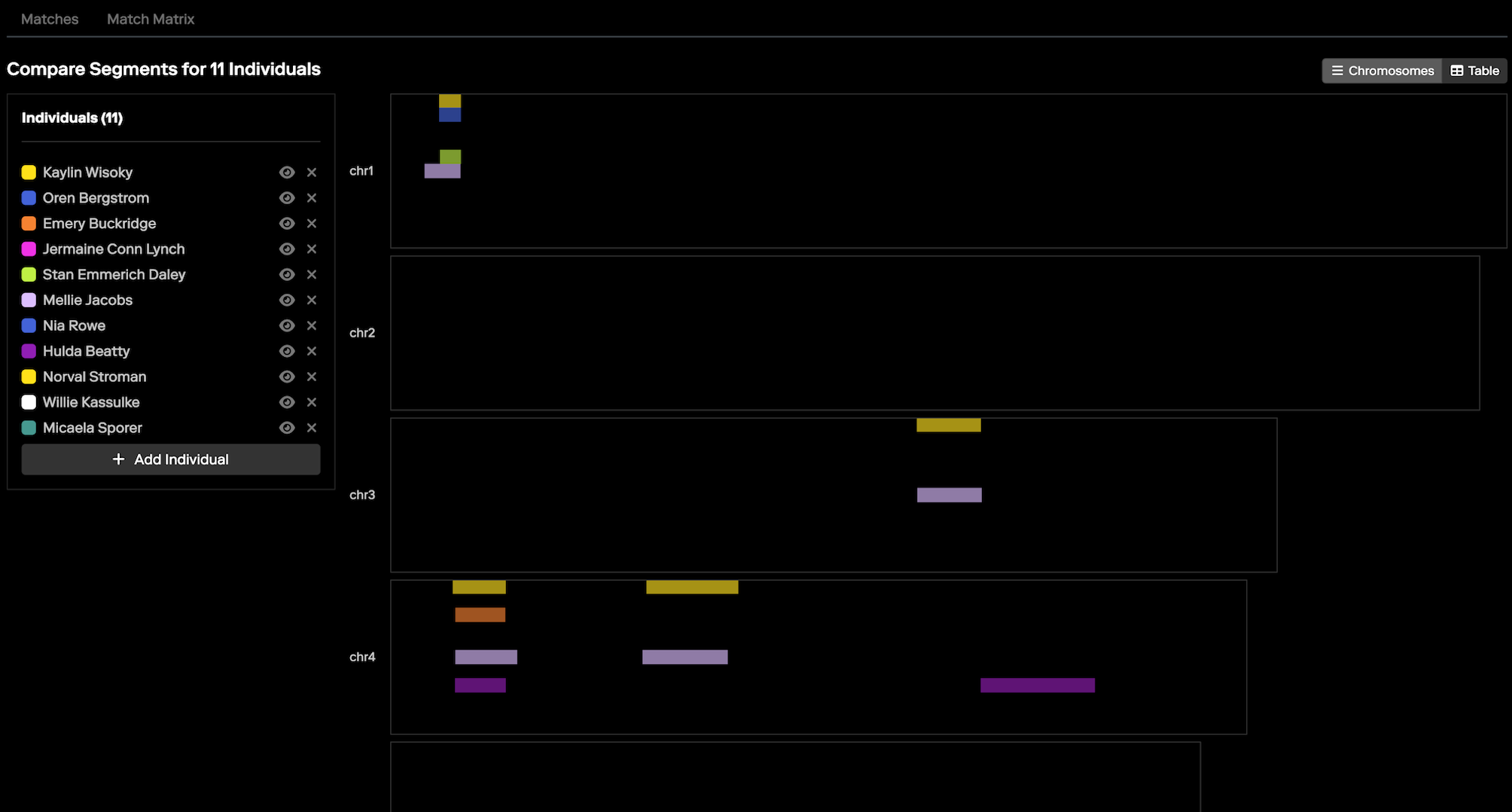
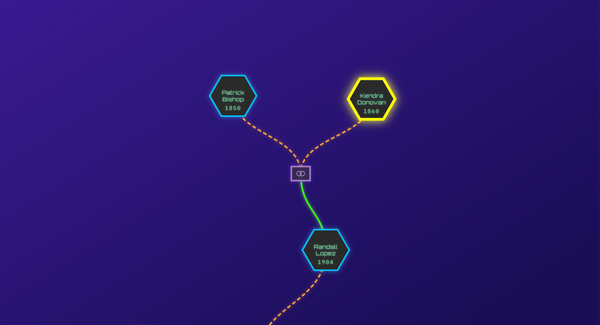
Once the relevant clusters are identified, the “Shared Matches” view can be used to further drill down and confirm the specific connections within those clusters. By using these tools in tandem, investigators can more effectively filter out irrelevant matches and focus on the data that will drive their investigation forward.
For example, suppose an investigator is working to identify unidentified human remains. After uploading the DNA profile to the MDFI platform, an investigator might find several clusters of matches, each pointing to different family lines. By focusing on the cluster with the strongest connections and most relevant biogeographical or ancestral context, the investigator can narrow down the list of potential identities more effectively. Clustering also helps in situations where distant matches are the only available leads, providing a structured way to interpret these connections and trace them back to a common ancestor.
Explore the New Features Today
We invite you to explore these new tools within the MDFI platform. Whether you’re working to identify a suspect or unknown remains, these features are here to support your efforts and make your life easier as you work to solve cases.
Let’s work together to unlock answers and bring justice to those who need it most. Get started here.