Labels carry weight—especially when it comes to identity. In the United States, the Census Bureau classifies race into categories like American Indian or Alaska Native, Asian, Black or African American, Native Hawaiian or Other Pacific Islander, and White, with a separate label for Latino ethnicity. However, these categories are often arbitrary social or political constructs. For instance, a Pashtun born in Pakistan is labeled “Asian,” while a Pashtun born in Afghanistan is considered “White” because the U.S. Census Bureau defines people of Middle Eastern descent as White. Afghanistan is grouped with the Middle East rather than South Asia, based on geopolitical boundaries rather than genetics.
These labels reflect the outcome of historical, social, and political processes. But what about scientific categories? In the world of personal genomics, consumer DNA testing companies use a different system. Their categories—such as European, Central & South Asian, East Asian, and Sub-Saharan African—are based on biogeography rather than geopolitical divisions. Though there are similarities with Census categories, the goal of consumer DNA testing companies is to classify people based on genetic clusters that align with geographic regions. Every category they use can be placed on a map, because genetic diversity across populations often reflects geographic separation over time.
The Genetic Foundation of Biogeographical Ancestry
The human genome is a vast landscape of 19,000 genes spread across 3 billion base pairs. Each region of the genome carries its own history, shaped by the complex processes of recombination and segregation that occur every generation. Biogeographical ancestry clustering attempts to reduce this complexity, distilling millions of genetic variations into a coherent narrative about where your ancestors came from.
On average, a person’s genome contains around 5 million variations from the human reference genome. While the vast majority of these variations are random mutations shared across all human populations, some reflect the unique history of specific populations. Over tens of thousands of years, as groups of people became isolated—like the Aboriginal Australians, who arrived in Australia 45,000 years ago—distinct genetic signatures emerged within these populations. Because of this long period of isolation, Aboriginal Australians developed a genetic profile that is easily distinguishable from that of other populations.
This process—population isolation and the development of unique genetic signatures—has occurred in many groups around the world. As a result, genetic variations that are shared within populations but rare outside of them can reveal a lot about an individual’s ancestral origins. These variations act as markers of biogeographical ancestry, and scientists have developed methods to extract this information rapidly and at scale.
Extracting Biogeographical Signals from DNA
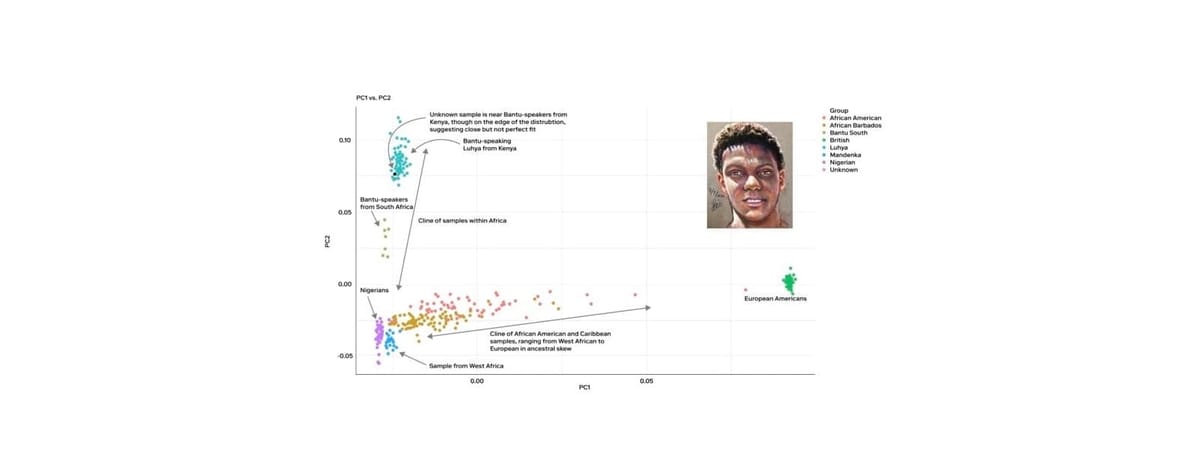
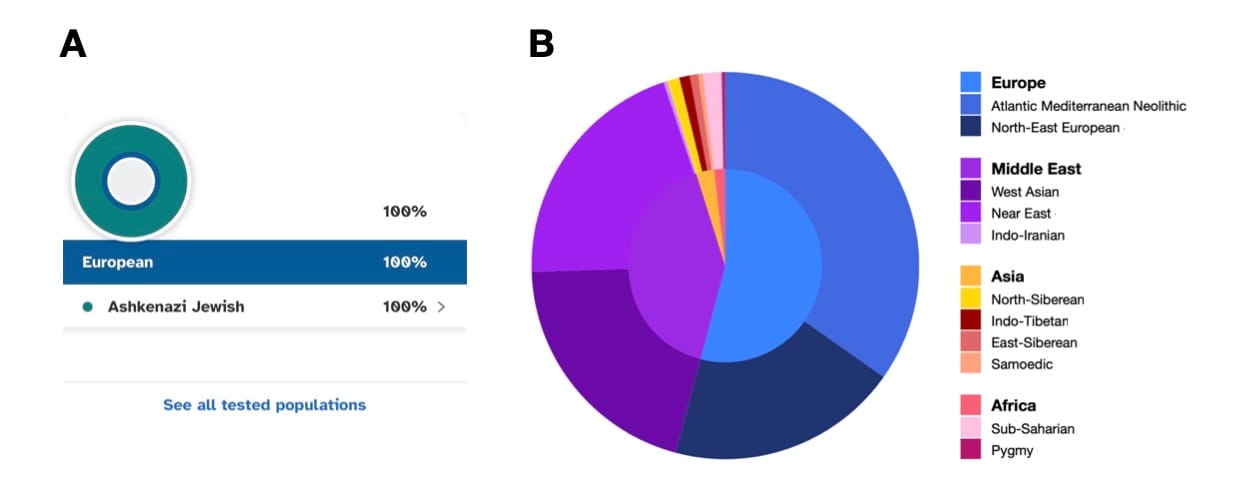
One of the most widely used methods for identifying biogeographical ancestry is principal component analysis (PCA). PCA takes complex genetic data and simplifies it into a few dimensions that can be plotted on a graph, revealing clusters that correspond to different populations. These dimensions, known as principal components, explain the most significant genetic variation in the data. The first dimension typically separates African populations from non-African populations, reflecting one of the deepest splits in human history. The second dimension often distinguishes West Eurasians from East Eurasians, capturing another major historical division.
By plotting individuals on a PCA graph, researchers can see how closely they align with these population clusters. PCA is particularly useful for visualizing genetic diversity and showing how different populations relate to each other across broad geographic regions.
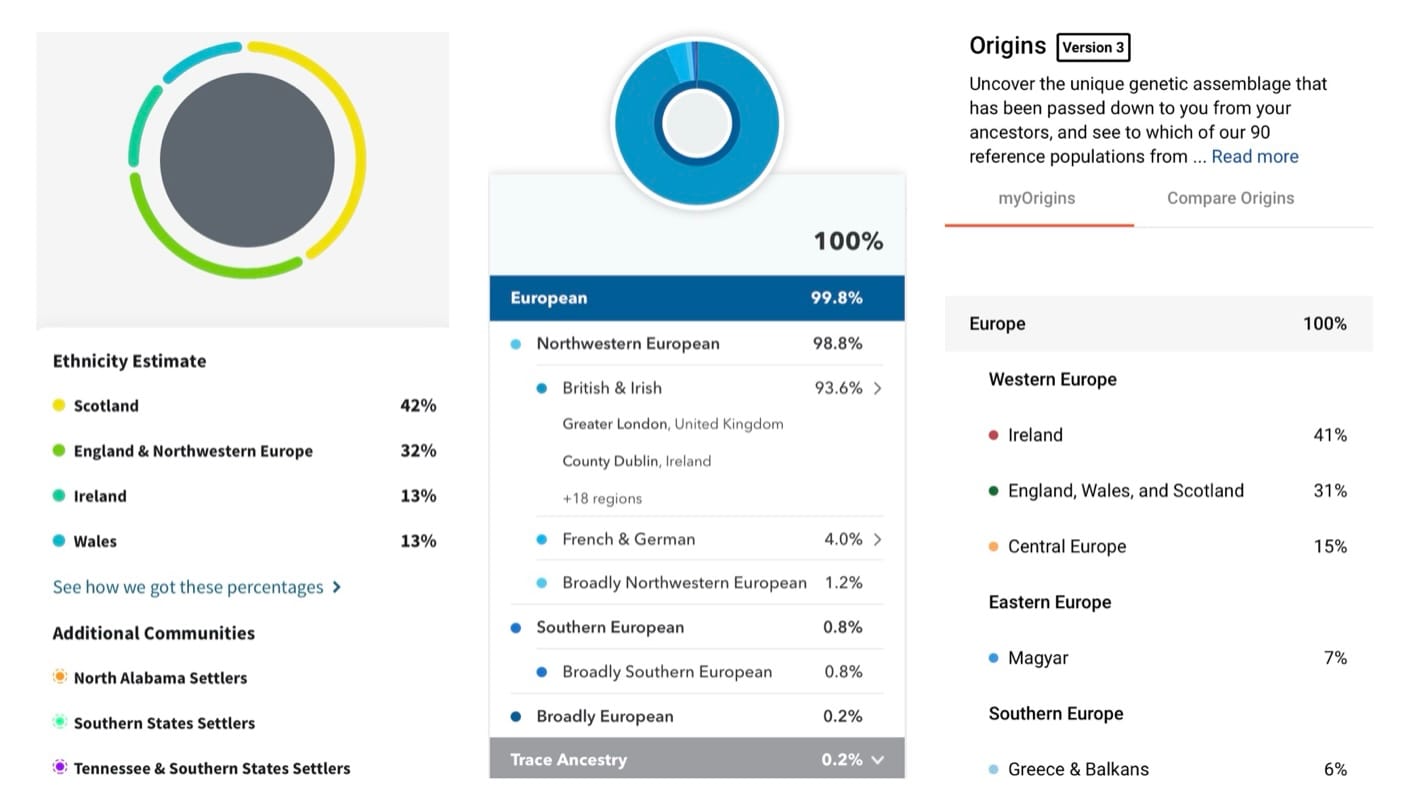
But sometimes, scientists use more targeted approaches that take prior information into account. For example, personal genomics companies often build specific ancestry categories based on the needs of their customers. In the U.S., many people have Jewish ancestry, so some companies have a separate category for identifying Jewish heritage. Unlike other groups, Jewish identity is not tied to a single geographic region. Instead, Jews have a long history of geographic mobility, but their shared genetic ancestry stems from their endogamous (in-group) marriage practices over centuries.
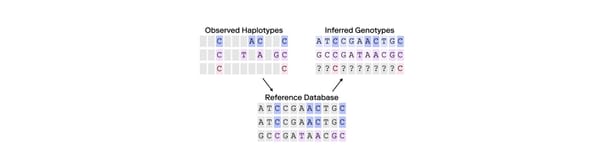
To detect Jewish ancestry—or the ancestry of other populations with unique histories—researchers can train their models on representative individuals from those populations. By comparing a customer’s DNA to a database of pre-selected individuals, they can determine if the customer shares genetic signatures specific to that population. For instance, a few hundred Jewish individuals are enough to capture much of the population-wide genetic variation within the Jewish community. The same is true for other populations like the Japanese. These methods can also identify people of admixed ancestry more accurately than PCA alone. For example, a person who is half-Jewish and half-Japanese might be positioned ambiguously on a PCA graph, but targeted methods would identify segments of DNA that match each population precisely.
The Importance of Labels in Forensic Genetic Genealogy
So, why do these labels matter? Biogeographical ancestry labels are one way we attempt to categorize the immense diversity in the human genome. They help simplify the complex genetic data we gather from DNA tests, turning it into something people can understand and relate to. While different companies may use slightly different labels or categories, the basic idea remains the same—these categories are designed to reflect real patterns of genetic diversity that have accumulated over time as populations have moved, migrated, and mixed.
It’s important to remember that all models are simplifications. Labels in genetic genealogy are useful, but they are not perfect. They are shaped by decisions about how to divide human populations into categories, and different platforms may make different choices based on their datasets and goals. However, these labels still provide valuable insights into our ancestry, allowing us to trace our genetic roots across the world.
In the context of forensic genetic genealogy, these labels can be powerful tools for identifying unknown individuals and tracing family lines. The ability to detect ancestral origins with increasing precision offers law enforcement and genealogists a way to narrow down the search for an unknown person’s identity.
Precision is the Promise of Forensic Genomics
With more affordable DNA testing, large datasets, and modern computing, we can supplement traditional forensic methods with new tools that will herald a future in which we not only produce more accurate measures of ancestry but also more specific information about a person’s origins, even to specific areas or population groups. Not only are these methods more specific, but biogeographical analysis methods are also reproducible and transparent. They rely on formal analysis and computational frameworks, not intuition and experience. As more data becomes available, they will also improve.
Let's look at a real case example. In May 2020, the body of a young Black man was discovered on the beach near Kahala Drive in Galveston, Texas. With no identifying information and no match in missing persons databases, the Galveston Police Department and Medical Examiner’s Office were unable to establish his identity, despite public outreach and the sharing of a forensic facial reconstruction. The case was logged in NamUs as UP69355 and soon went cold.
Later that year, the Galveston Police partnered with Othram and we built a forensic DNA profile using Forensic-Grade Genome Sequencing®. Investigators initially thought that the man was African American. Our analysis immediately revealed that the man’s biogeographical ancestry was actually substantially East African and that he might be Somali Bantu or Kenyan Bantu. He didn't have European and West African ancestry, which would be expected from an African American person. This new information prompted investigators to consider that he may have been visiting from overseas. Further investigation confirmed the young man was a 24-year-old exchange student from Tanzania.
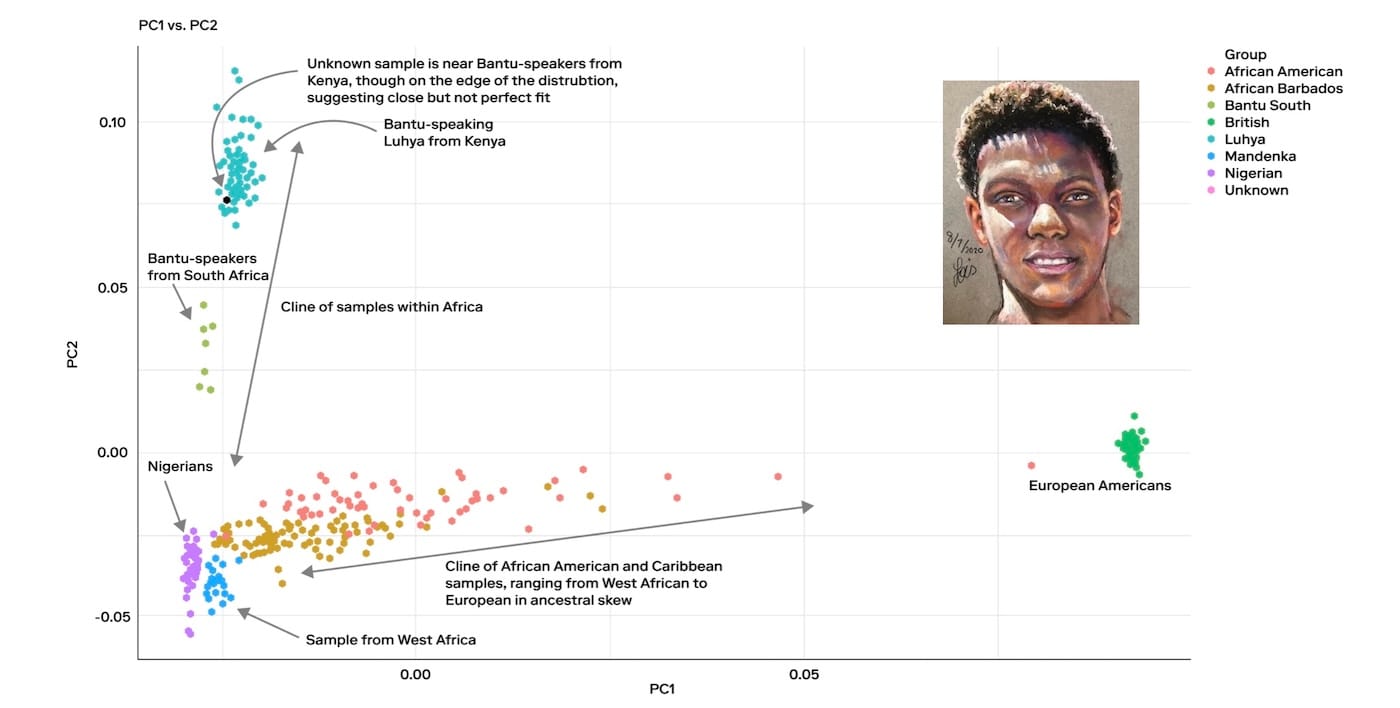
In this case, knowing the correct (and precise) biogeographical ancestry was key to helping investigators identify this young man. When working cold cases, you should expect, and in fact demand, this complete genomic analysis because every detail matters and any detail could be a clue.
Conclusion: Every Detail Matters and Any Detail Could be a Clue
If you are not getting a complete analysis from your vendors or internal methods, or if you are not ready to onboard this new technology in your own forensic setting yet, come to Othram. Your investigation can't afford to miss a key detail or fact. Evidence is finite and if you use the wrong methods or don't collect enough data to perform a complete analysis, the case can be lost forever.
Our team operates the world's first purpose-built forensic laboratory for forensic genetic genealogy. We developed Forensic-Grade Genome Sequencing® or FGGS® to enable ultra-sensitive detection of distant relationships. It's part of our Multi Dimensional Forensic Intelligence (MDFI) platform.
More forensic genetic genealogy cases have been solved with Othram FGGS® than any other method. Let’s work together to unlock answers and bring justice to those who need it most. Get started here.